📢 Chen et al. recently reported H3-OPT, a model for predicting structures of the heavy chain 3 of antibody (Ab) complementarity-determining regions (CDR-H3) based on AlphaFold2 (AF2). For both monoclonal Abs (having both heavy and light chains) and nanobodies (with a single-domain heavy chain), the CDR-H3 loop plays a key role in antigen binding and is thus the most diverse Ab region in terms of amino acid length and composition.
💉 Given the importance of the CDR-H3, accurate development of Ab-based therapeutics is dependent on experimental structures of candidate Abs, a process which is bottlenecked by both cost and time. Thus, accurate computational prediction methods would facilitate the therapeutic Ab development pipeline. However, existing methods struggle to generate high quality predictions of the CDR-H3 loop regions because of the inherent challenge of predicting loop structures.
🔥 H3-OPT integrates the strengths of AF2 with a pre-trained protein language model (PLM) to predict CDR-H3 structures. Based on the observation that AF2-predicted Ab structures show an overall high quality, H3-OPT extracts the structural features and uses the information to generate refined CDR-H3 structures.
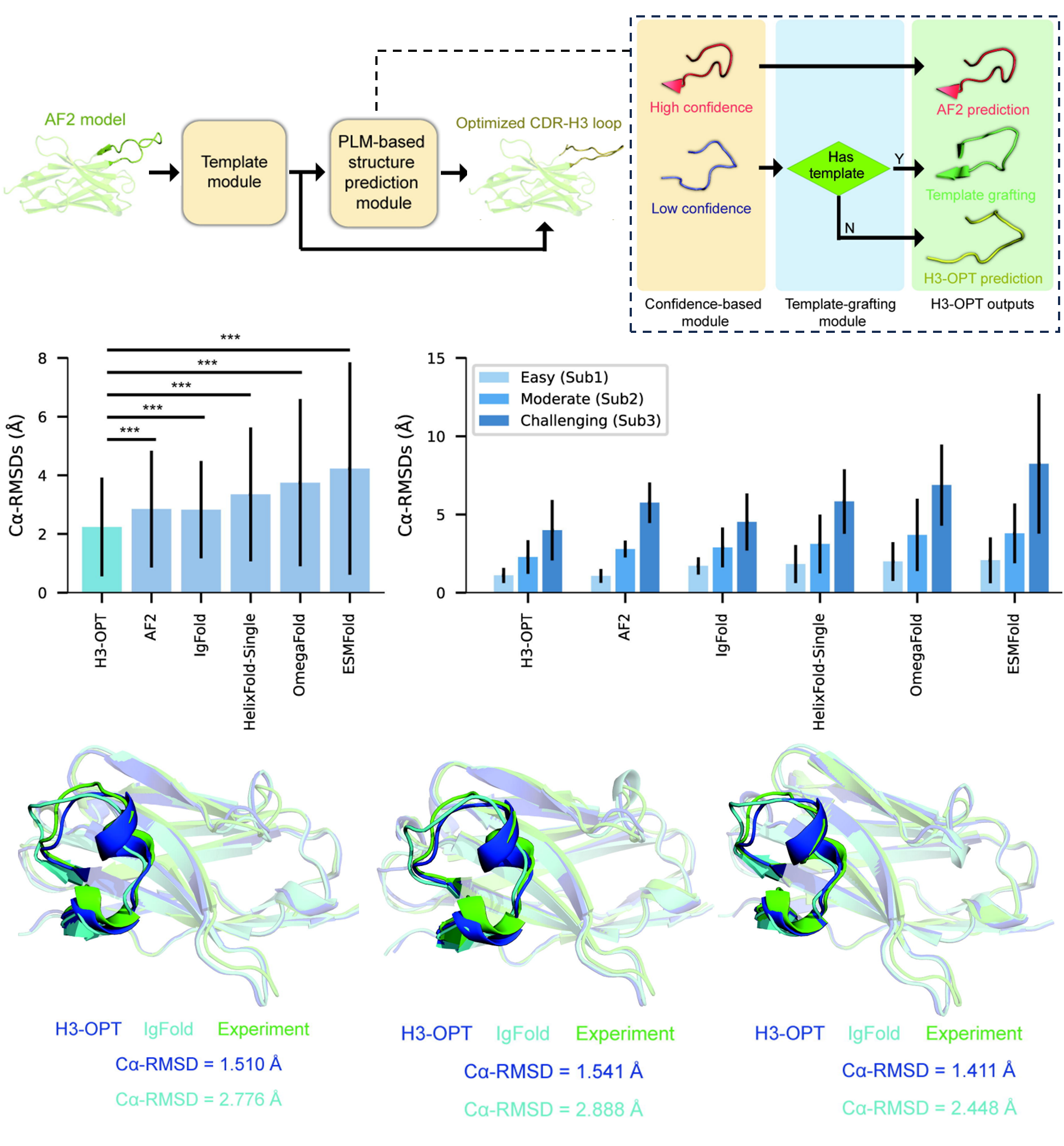
H3-OPT consists of two modules: a template module and a PLM-based structure prediction module (PSPM). The template module dictates whether the PSPM is deployed for a particular query. The PSPM is further comprised of two sub-modules: a confidence-based module that evaluates the quality of the CDR-H3 from AF2, and a template-grafting module which identifies a suitable PDB template to replace low quality AF2 H3 loop.
🔢 On a benchmark dataset, H3-OPT outperforms other state-of-the-art methods, including AF2, IgFold, HelixFold-Single, ESMFold, and OmegaFold, by achieving an RMSD (to experimental structure) of 2.24 Å, compared to 2.85 Å and 2.87 Å for the next best methods, AF2 and IgFold, respectively.
Furthermore, H3-OPT was evaluated using recently deposited PDB structures of three anti-VEGF variants, giving predictions with RMSDs of 1.510 Å, 1.541 Å, and 1.411 Å for the three variants. Predictions by IgFold, however, gave RMSDs of 2.776 Å, 2.888 Å, and 2.448 Å.
💪 H3-OPT also exhibited improved performance on other tasks, such as the prediction of Ab CDR-H3 surface properties (including surface amino acids, solvent-accessible surface areas, and surface charge distribution) and antibody-antigen interactions.
Open Questions
- Improving Predictive Power:
- How can H3-OPT be optimized to handle the most challenging CDR-H3 loops found in rare or unconventional antibodies?
- Can incorporating additional data from single-cell sequencing and high-throughput mutagenesis studies further refine H3-OPT’s predictions?
- Synergy with Experimental Approaches:
- What innovative strategies can be developed to integrate H3-OPT predictions with cryo-EM or NMR spectroscopy data for real-time structural validation?
- How might H3-OPT predictions be used to guide experimental design in directed evolution campaigns for therapeutic antibodies?
- Applications in Biologics Development:
- How can H3-OPT be utilized to streamline the design of next-generation bispecific and multispecific antibodies?
- What role could H3-OPT play in accelerating the development of antibody-drug conjugates (ADCs) by predicting optimal binding and linker sites?
- Diverse Antibody Repertoires:
- How does H3-OPT perform when applied to antibodies derived from unconventional sources, such as shark VNARs or camelid VHHs?
- Can H3-OPT be adapted to predict the structures of synthetic antibody libraries, potentially expanding the diversity of therapeutic candidates?
- Enhancing User Experience:
- What user-friendly interfaces and visualization tools can be developed to make H3-OPT accessible to a broader range of researchers, including those with limited computational backgrounds?
- How can H3-OPT be integrated into existing biologics discovery platforms to create seamless workflows from prediction to experimental validation?
- Technical Innovations:
- How can the computational demands of H3-OPT be reduced to allow for rapid, large-scale antibody screening projects?
- What novel machine learning techniques could be applied to improve H3-OPT’s accuracy and speed, particularly for antibodies with large or complex epitopes?
Conclusion
H3-OPT stands out as an effective method for overcoming the challenges associated with predicting the structure of antibody CDR-H3 regions. By integrating the robust capabilities of AlphaFold2 with a protein language model, H3-OPT achieves higher accuracy and reliability in its predictions. Its modular design allows for flexible adaptation based on the quality of initial predictions, making it a valuable tool in the field of antibody development.
Resources
- Paper: H3-OPT: Accurate prediction of CDR-H3 loop structures of antibodies with deep learning
- GitHub: H3-OPT
